Source: blogs.plos.org
Author: Catherine Chang et al.
Turning red after consuming alcohol may seem like a mere social inconvenience. Yet, behind this red complexion lies a far more serious problem. ALDH2 deficiency, more commonly known as Alcohol Flushing Syndrome or Asian Glow, is a genetic condition that interferes with the metabolism of alcohol. As a result, people with ALDH2 deficiency have increased risks of developing esophageal and head and neck cancers . Globally, this deficiency affects 540 million people — 8% of the world population. In East Asia (which includes Japan, China, and Korea), this is a much bigger problem, where 36% of the population is affected [1]. In our home, Taiwan, approximately 47% of the population carries this genetic mutation — the highest percentage in the world [2]!
Normally, ethanol is first converted to acetaldehyde (a toxic intermediate) by the enzyme alcohol dehydrogenase (ADH). A second enzyme, aldehyde dehydrogenase 2 (ALDH2), then converts toxic acetaldehyde into acetate, a compound which can be safely metabolized in the body. For people who carry wild type ALDH2*1, acetaldehyde can be broken down quickly. People with ALDH2 deficiency, however, have a point mutation which leads to the less efficient mutant ALDH2*2 [3], [4]. Enzymatic activity in ALDH2-deficient individuals can be as low as 4% compared to wild type [4], [5], [6], [7]. As a result, acetaldehyde accumulates and induces an inflammatory response that causes the skin to flush after drinking alcohol [8]. Turning red is the most obvious result of ALDH2 deficiency, but symptoms also include headaches, dizziness, hypotension, and heart palpitations [5], [9].
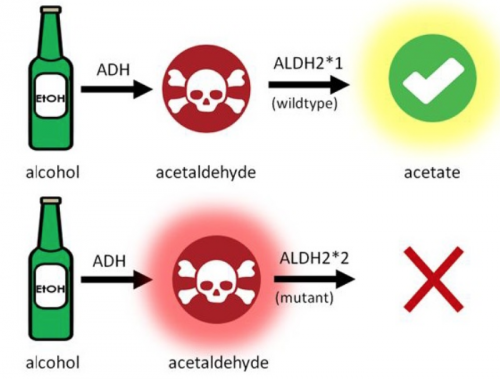
Acetaldehyde accumulates in ALDH2-deficient individuals. Ethanol is first converted to a toxic intermediate, acetaldehyde, by ADH, then converted to acetate by wild type ALDH2*1. The mutant form, ALDH2*2, cannot fully convert acetaldehyde into acetate, and toxic acetaldehyde accumulates as a result.
For people who are ALDH2-deficient and drink, acetaldehyde can accumulate to toxic levels. The International Agency for Research on Cancer classifies acetaldehyde associated with alcohol consumption as a Group 1 carcinogen [10]. Acetaldehyde levels over 50 μM are considered toxic and cause mutations in DNA, and studies show that the strongest effects are seen in the mouth [11], [12]. After consuming roughly 2 to 3 servings of alcohol (0.5-0.6 g alcohol/kg body weight), salivary acetaldehyde levels in ALDH2-deficient individuals reached over 100 μM, compared to normal levels of <20 μM without drinking [13], [14], [15], [16]. Because of the increased salivary acetaldehyde, people with ALDH2 deficiency are 2 to 8 times more likely to develop head and neck cancers (including oral cancer, pharyngeal cancer, laryngeal cancer, etc.), and 2 to 12 times more likely to develop esophageal cancer compared to people with normal ALDH2*1 [17-25].
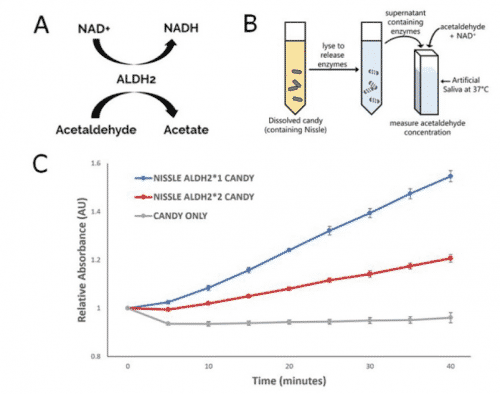
Our ALDH2*1 probiotic candy significantly reduces acetaldehyde levels in simulated oral conditions. (A) The conversion of acetaldehyde to acetate by ALDH2 uses NAD+ and produces NADH. (B) Experimental setup. The candies were dissolved, the probiotic (Nissle) was lysed to release ALDH2 enzymes, and the supernatant was placed into artificial saliva. NADH concentration was measured by taking absorbance readings at 340 nm. (C) Enzymatic activity of ALDH2*1 and ALDH2*2 from the probiotic candies. A negative control of candy without Nissle was also included (gray). Under these conditions, the ALDH2*1 candies metabolized significantly more acetaldehyde compared to both the ALDH2*2 candies and the negative control. Error bars represent standard error.
To directly address the increased esophageal and head and neck cancer risks, we developed a probiotic (E. coli Nissle 1917) candy carrying recombinant human ALDH2*1 to maintain normal acetaldehyde levels in the mouths of ALDH2-deficient individuals. We tested the candy’s ability to break down acetaldehyde by measuring NADH, a byproduct of acetaldehyde metabolism. In simulated oral conditions, we observed a significant decrease in acetaldehyde levels when we added the contents of our ALDH2*1 candy (compared to the mutant ALDH2*2 or control candy). Through mathematical modeling, we also determined the exact amount of recombinant ALDH2*1 needed in each piece of candy. Our modeling shows that if a consumer eats our candy while drinking, the released ALDH2*1 will be able to combat the high salivary acetaldehyde levels and match the normally low levels found in wild type individuals.
Nearly half of Taiwan’s population is ALDH2 deficient. To combat the increased cancer risks associated with this deficiency, we developed and tested a method to regulate acetaldehyde levels in ALDH2-deficient individuals.
The TAS_Taipei iGEM Team have produced a full research article detailing their project. You can access that article here.
Note:Please note that the team’s full research article has not been peer-reviewed.
Authors: Catherine Chang, Tim Ho, Iris Huang, Justin Wu
References:
1. Brooks PJ, Enoch MA, Goldman D, Li TK, Yokoyama A. (2009). The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 24;6(3):e50.
2. Chang JS, Hsiao JR, Chen CH. (2017). ALDH2 polymorphism and alcohol-related cancers in Asians: a public health perspective. J Biomed Sci. 24(1):19.
3. Larson HN, Weiner H, Hurley TD. (2005). Disruption of the Coenzyme Binding Site and Dimer Interface Revealed in the Crystal Structure of Mitochondrial Aldehyde Dehydrogenase “Asian” Variant. J Biol Chem. 280(34):30550-6.
4. Farrés J, Wang X, Takahashi K, Cunningham SJ, Wang TT, Weiner H. (1994). Effects of changing glutamate 487 to lysine in rat and human liver mitochondrial aldehyde dehydrogenase. A model to study human (Oriental type) class 2 aldehyde dehydrogenase. J Biol Chem. 13;269(19):13854-60.
5. Chen CH, Ferreira JCB, Gross ER, Mochly-Rosen D. (2014). Targeting Aldehyde Dehydrogenase 2: New Therapeutic Opportunities. Physiol Rev. 94(1):1-34.
6. Zhou J, Weiner H. (2000). Basis for half-of-the-site reactivity and the dominance of the K487 oriental subunit over the E487 subunit in heterotetrameric human liver mitochondrial aldehyde dehydrogenase. Biochemistry. 39(39):12019-24.
7. Gross ER, Zambelli VO, Small BA, Ferreira JCB, Chen CH, Mochly-Rosen D. (2015). A personalized medicine approach for Asian Americans with the aldehyde dehydrogenase 2*2 variant. Annu Rev Pharmacol Toxicol. 55:107-27.
8. Ijiri I. (1999). [Biological actions of acetaldehyde]. Nihon Hoigaku Zasshi. 53(3):285-95.
9. Lai CL, Yao CT, Chau GY, Yang LF, Kuo TY, Chiang CP, Yin SJ. (2014) Dominance of the inactive Asian variant over activity and protein contents of mitochondrial aldehyde dehydrogenase 2 in human liver. Alcohol Clin Exp Res. 2014 Jan;38(1):44-50.
10. International Agency for Research on Cancer. (n.d.). List of Classifications, Volumes 1-122. Retrieved from https://monographs.iarc.fr/list-of-classifications-volumes/ .
11. Yamaguchi H, Hosoya M, Shimoyama T, Takahashi S, Zhang JF, Tsutsumi E, Suzuki Y, Suwa Y, Nakayama T. (2012). Catalytic removal of acetaldehyde in saliva by a Gluconobacter strain. J Biosci Bioeng. 114(3):268-74.
12. Kocaelli H, Apaydin A, Aydil B, Ayhan M, Karadeniz A, Ozel S, Yilmaz E, Akgün B, Eren B. (2014) Evaluation of potential salivary acetaldehyde production from ethanol in oral cancer patients and healthy subjects. Hippokratia. 18(3): 269–274.
13. Homann N, Jousimies-Somer H, Jokelainen K, Heine R, Salaspuro M. (1997a). High acetaldehyde levels in saliva after ethanol consumption: methodological aspects and pathogenetic implications. Carcinogenesis. 18(9):1739-43.
14. Yokoyama A, Tsutsumi E, Imazeki H, Suwa Y, Nakamura C, Mizukami T, Yokoyama
(2008). Salivary acetaldehyde concentration according to alcoholic beverage consumed and aldehyde dehydrogenase-2 genotype. Alcohol Clin Exp Res. 32(9):1607-14.
15. Lachenmeier DW, Monakhova YB. (2011). Short-term salivary acetaldehyde increase due to direct exposure to alcoholic beverages as an additional cancer risk factor beyond ethanol metabolism. J Exp Clin Cancer Res. 30 (3).
16. Stornetta A, Guidolin V, Balbo S. (2018). Alcohol-Derived Acetaldehyde Exposure in the Oral Cavity. Cancers (Basel). 10(1). pii: E20.
17. Chao YC, Wang LS, Hsieh TY, Chu CW, Chang FY, Chu HC. (2000). Chinese Alcoholic Patients with Esophageal Cancer are Genetically Different from Alcoholics with Acute Pancreatitis and Liver Cirrhosis. Am J Gastroenterol. 95(10):2958-2964.
18. Yang SJ, Wang HY, Li XQ, Du HZ, Zheng CJ, Chen HG, Mu XY, Yang CX. (2007). Genetic Polymorphisms of ADH2 and ALDH2 Associated with Esophageal Cancer Risk in Southwest China. World J Gastroenterol. 13(43):5760-5764.
19. Yokoyama A, Muramatsu T, Omori T, Yokoyama T, Matsushita S, Higuchi S, Maruyama K, Ishii H. (2001). Alcohol and Aldehyde Dehydrogenase Gene Polymorphisms and Oropharyngolaryngeal, Esophageal and Stomach Cancers in Japanese Alcoholics. Carcinogenesis. 22(3): 433-439.
20. Matsuo K, Hamajima N, Shinoda M, Hatooka S, Inoue M, Takezaki T, Tajima K. (2001). Gene-Environment Interaction between an Aldehyde Dehydrogenase-2 (ALDH2) Polymorphism and Alcohol Consumption for the Risk of Esophageal cancer. Carcinogenesis. 22(6): 913-916.
21. Cui R, Kamatani Y, Takahashi A, Usami M, Hosono N, Kawaguchi T, Tsunoda T, Kamatani N, Kubo M, Nakamura Y, Matsuda K. (2009). Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology. 2009 Nov;137(5):1768-75.
22. Lee CH, Lee JM, Wu DC, Goan YG, Chou SH, Wu IC, Kao EL, Chan TF, Huang MC, Chen PS, Lee CY, Huang CT, Huang HL, Hu CY, Hung YH, Wu MT. (2007). Carcinogenetic impact of ADH1B and ALDH2 genes on squamous cell carcinoma risk of the esophagus with regard to the consumption of alcohol, tobacco and betel quid. Int J Cancer. 122(6):1347-56.
23. Wu M, Chang SC, Kampman E, Yang J, Wang XS, Gu XP, Han RQ, Liu AM, Wallar G, Zhou JY, Kok FJ, Zhao JK, Zhang ZF. (2013). Single nucleotide polymorphisms of ADH1B, ADH1C and ALDH2 genes and esophageal cancer: a population-based case-control study in China. Int J Cancer. 132(8):1868-77.
24. Huang CC, Hsiao JR, Lee WT, Lee YC, Ou CY, Chang CC, Lu YC, Huang JS, Wong TY, Chen KC, Tsai ST, Fang SY, Wu JL, Wu YH, Hsueh WT, Yen CJ, Wu SY, Chang JY, Lin CL, Wang YH, Weng YL, Yang HC, Chen YS, Chang JS. (2017). Investigating the Association between Alcohol and Risk of Head and Neck Cancer in Taiwan. Sci Rep. 7(1):9701.
25. Hiraki A, Matsuo K, Wakai K, Suzuki T, Hasegawa Y, Tajima K. (2007). Gene–gene and gene–environment interactions between alcohol drinking habit and polymorphisms in alcohol-metabolizing enzyme genes and the risk of head and neck cancer in Japan. Cancer Science. 98: 1087-1091.


Leave A Comment
You must be logged in to post a comment.